A recent study, “Comparative proteomic landscapes elucidate human preimplantation development and failure”, published in Cell on January 24. Research teams led by Liu Zhen, Sun Yidi, and Zhu Wencheng from the Center for Excellence in Brain Science and Intelligence Technology of the Chinese Academy of Sciences, in collaboration with Mu Liangshan from the Reproductive Medicine Center, Zhongshan Hospital, Fudan University, and Li Chen at the Center for Single-Cell Omics, School of Public Health, Shanghai Jiao Tong University School of Medicine, uncovered the success and failure of early human embryonic development from the protein dynamic level.

Mammalian preimplantation embryo development includes key biological events such as fertilization, zygotic genome activation (ZGA), and differentiation of trophoblast cells and inner cell clusters (first lineage differentiation). Benefiting from the continuous development of single-cell and low-input multi-omics technology, previous studies have shown that there is a large amount of uncoupling regulation between transcription and translation in early embryonic development [1-4]. However, understanding mammalian preimplantation development, particularly in humans, at the proteomic level remains limited [5-7].
The researchers first improved the previously developed CS-UPT hypersensitive proteome technology system [8, 9]. Then, the cross-species, in-depth proteome landscape of preimplantation embryo development, including nearly 8,000 proteins in humans and over 6,300 proteins in mice, was drawn (Figure 1).
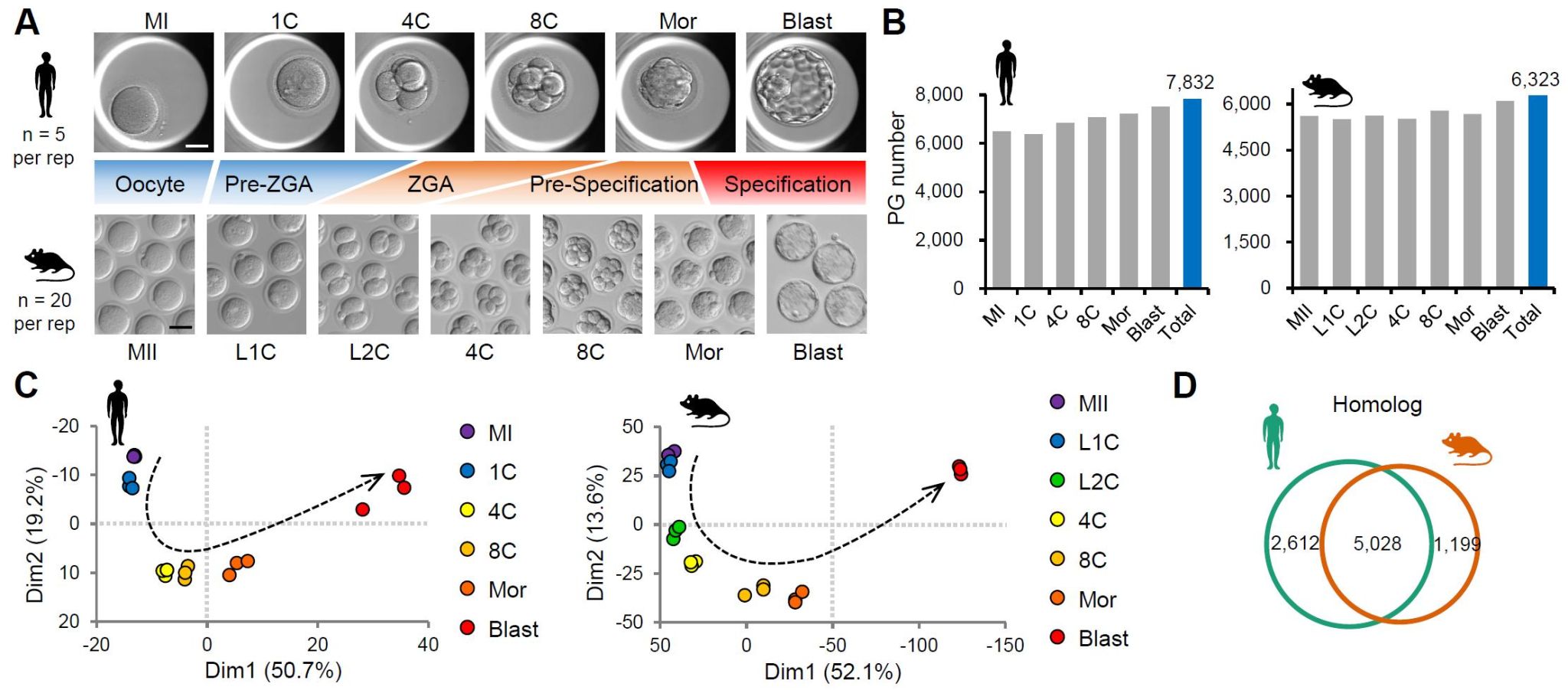
Through cross-species comparisons, the researchers found that the differences in protein dynamics between humans and mice were mainly concentrated before and after zygotic genome activation (pre-ZGA and ZGA). This finding provides a proteomic basis for understanding the regulation of differences in zygotic genomes between species. After integrated analysis with the previous translatome data [1, 3], the study showed that compared with the translational dynamics, proteome dynamics can be divided into three major patterns: consistent, delayed and stable. The researchers further validated the observed divergent dynamics between proteins and mRNA translation by conducting translation inhibition and protein degradation inhibition experiments druing maternal-to-zygotic transition (MZT) in mice.
ZGA genes are generally defined by their transcriptional activation at a certain degree when comparing to early zygotes or oocytes. However, how these genes continue to express after ZGA and the fate of their transcripts remain elusive. Using the recent transcriptomic and translatomic datasets, two mainly types, increasing ZGA (I-ZGA) and transient ZGA (T-ZGA), were defined. Interestingly, the researchers found that proteins produced by many T-ZGA transcripts accumulate continuously after transcriptional and translational activation, reaching their highest abundance during the first lineage differentiation. All of these were termed as “ZGA-burst proteins” and likely in preparation for the first lineage differentiation. The researchers selected representative transcription factors and designed functional experiments to validate their important and novel roles in early embryonic development.
Finally, the researchers explored the potential of human proteomic landscape as a reference in understanding the formation of poor quality (PQ) embryos [10]. The researchers recruited more than 100 patient-couples, and performed single-embryo proteomic (SEP) analyses of more than 140 PQ embryos and 9 good quality (GQ) embryos. On average, approximately 4,800 proteins in each embryo, and a total of nearly 6,000 proteins were identified. Unsupervised clustering analysis revealed a distinct proteomic profile for PQ embryos compared to GQ embryos, identifying three subgroups within the PQ embryos. What’s more, PQ embryos from the same patient-couples exhibited proteomic differences, while GQ embryos remained similar. In addition to dysregulation of maternal protein degradation and increase in newly synthesized proteins, these PQ embryos generally exhibit dysregulation of “stable” proteins. The study also revealed a number of candidate proteins that may lead to the formation of PQ embryos.

In conclusion, this study integrates technology improvement, resource mining, discovery exploration, and clinical application, providing new perspectives, resources, and ideas for understanding preimplantation embryonic development in humans and mice, as well as human preimplantation development failures.
References:
1. Xiong, Z., et al., Ultrasensitive Ribo-seq reveals translational landscapes during mammalian oocyte-to-embryo transition and pre-implantation development. Nat Cell Biol, 2022. 24(6): p. 968-980.
2. Zhang, C., et al., Profiling and functional characterization of maternal mRNA translation during mouse maternal-to-zygotic transition. Sci. Adv., 2022. 8(5): p. eabj3967.
3. Zou, Z., et al., Translatome and transcriptome co-profiling reveals a role of TPRXs in human zygotic genome activation. Science, 2022. 378(6615): p. abo7923.
4. Ozadam, H., et al., Single-cell quantification of ribosome occupancy in early mouse development. Nature, 2023. 618(7967): p. 1057-1064.
5. Gao, Y., et al., Protein Expression Landscape of Mouse Embryos during Pre-implantation Development. Cell Rep., 2017. 21(13): p. 3957-3969.
6. Dang, Y., et al., Functional profiling of stage-specific proteome and translational transition across human pre-implantation embryo development at a single-cell resolution. Cell Discov, 2023. 9(1): p. 10.
7. Israel, S., et al., An integrated genome-wide multi-omics analysis of gene expression dynamics in the preimplantation mouse embryo. Sci Rep, 2019. 9(1): p. 13356.
8. Gu, L., et al., Ultrasensitive proteomics depicted an in-depth landscape for the very early stage of mouse maternal-to-zygotic transition. J Pharm Anal, 2023. 13(8): p. 942-954.
9. Zhu, W., et al., Reading and writing of mRNA m6A modification orchestrate maternal-to-zygotic transition in mice. Genome Biol, 2023. 24(1): p. 67.
10. Li, Y., et al., Transcriptomic profiling of day 3 human embryos of poor quality reveals molecular links to divergent developmental trajectories. Cell Rep, 2024. 43(11): p. 114888.




